Lab's projects
LAGM's lines of research are done in an interdisciplinary manner. Our projects involve collaborations with agronomists, biochemists, physiologists, computer scientists, statisticians, botanists. The lab's research lines focus on the following overlapping areas:
Biotechnology Applied in the Study of Enzymes that Acted in the Degradation of Plant Biomass
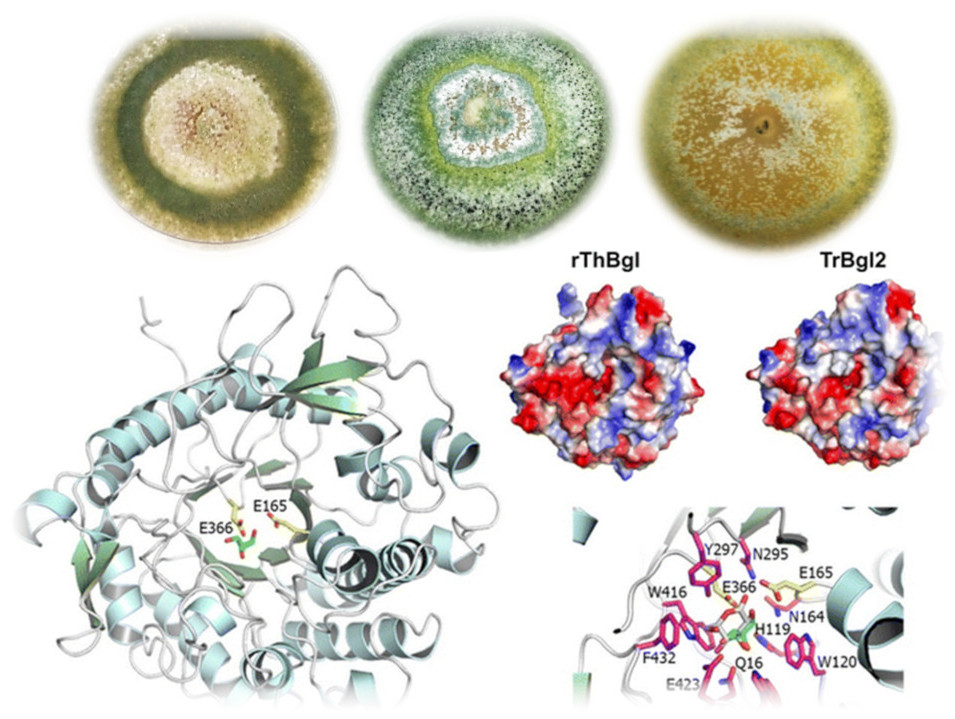
Fungi of the genus Trichoderma are of great biotechnological relevance due to their capacity to produce enzymes that act in the degradation of vegetal biomass, this potential can be exploited for the production of biofuels, with special attention in Brazil for second generation bioethanol. In this context, the research group related to Trichoderma has as main objective to carry out genetic studies of different species of this genus, including: T. harzianum, T. reesei and T. atroviride, aiming to establish the genetic and molecular bases related to the efficacy of metabolism of carbohydrates. For this, data from different approaches are integrated seeking a broad understanding of the process of degradation, among which stands out: genomics with the prospecting of genes and regulatory regions; analysis of global gene expression under biomass degradation conditions; proteomics with analysis of secreted enzymes; heterologous expression of proteins for biochemical studies and protein engineering analyzes looking more efficient enzymes for the industrial application. As a result of our research, new enzymes may be used during the enzymatic hydrolysis process, thus making the production of bioethanol more efficient and economically viable.
Evolution and conservation of Brazilian native plant species
Considering the megabiodiversity of Brazil, it is not surprising that a number of plant species are useful and have economic and cultural importance in regional and global scales. Among the Brazilian biomes, Amazonia is not only a reservoir of biological diversity, but also it is one of the major centers of plant domestication in the world. In this research area, we aim to characterize the genetic diversity of Amazonian crops based on the variation detected through molecular markers. Using population genetics and genomics approaches, we aim to infer about the evolutionary processes that occurred in Amazonian crops. These same approaches are also applied for the evaluation of the genetic diversity of native plant species from other hotspots of biodiversity, such as the Cerrado and the Atlantic Forest, aiming to promote strategies for their management and conservation. The comprehension of genetic variability and structuring of populations is essential to understand the evolutionary history, as well as for the conservation of the genetic resources of native Brazilian plant species.
Comparative evolutionary studies in mangrove plants species: patterns of natural selection, population structure, and biogeographic history of the community
Mangroves are ecosystems that occur at the border between the sea and the continent in tropical and subtropical latitudes, which play important ecological roles and serve as a transitional or permanent habitat for an extremely diverse fauna. However, urban, industrial, agricultural, and aquaculture, coupled with the recent accelerated process of global climate change (GCM), threaten the future of these ecosystems and the consequences are difficult to predict. Our research group seeks to describe the phylogeographic history and the roles of natural selection, dispersal processes, and demographic factors in the organization of the genetic diversity of plant species widely distributed throughout the Atlantic and East Pacific mangroves. Through the application of microsatellite and single nucleotide polymorphism (SNPs) data, our work seeks to contribute to the prediction of responses of mangrove communities to climate and to define strategies for the management, preservation, and recovery of the most vulnerable mangroves.
Genetics and Genomics of Tropical Forage Grasses
Tropical forage grasses are the basis for livestock feed and play a fundamental role in the Brazilian economy. Due to inadequate management conditions and biotic / abiotic factors, we have a scenario of extensive areas of degraded pasture, while in other areas it is believed that pasture productivity is less than its estimated capacity. In this context, it is necessary to obtain cultivars with high productivity and adapted to adverse environmental conditions, in order to maintain and overcome the present levels of productivity in tropical conditions. The main forage grasses grown in Brazil, exotic (Urochloa spp. or Megathyrsus spp.) or natives (Paspalum spp.), present complex genomes (polyploids), are apomictic, and there are few genetic and genomic resources available. Such a lack of knowledge limits the progress in obtaining hybrids and improved varieties, both in quantity and in quality by breeding programs. Aware of this situation, our research group has been working since 2005 on the development of genetic and genomic studies of the main tropical forages, aiming to generate molecular tools such as molecular markers (SSR and SNP), transcriptomes and genetic maps, in order to increase understanding of genetic variability and to help genetic breeding programs.
Sugarcane Transcriptomics and Genomics
Sugarcane (Saccharum ssp.) is among the main crops grown in Brazil due its derivatives: sugar and ethanol fuel. The modern sugarcane cultivar is an interspecific hybrid between S. officinarum L. (2n = 80) and S. spontaneum L. (2n = 40-128), with high ploidy level and aneuploidy, resulting in a very complex genome. The projects related to sugarcane aim to understand the genome structure and genome functioning, especially the genetic and genomic mechanisms linked to gene expression and gene regulation as well the relationship between ploidy level and allelic dosage. The knowledge The objectives also include applying these knowledges in the search for target genes reaching a sugarcane improvement, through the investigation of genomic regions related to sugar accumulation, disease resistance, such as Sugarcane yellow leaf virus.
Rubber tree
Rubber tree (Hevea brasiliensis) is native to the Amazon rain forest and the main source of natural rubber in the world, a raw material of great importance in several industry sectors. The need for new rubber clones adapted to different climatic regions, known as escape regions, is an important point for the success of the heveiculture in Brazil and in the world. The traditional plant breeding cycle for rubber tree can take approximately 30 years to be concluded. Therefore, it is fundamental to develop new techniques of early evaluation, which make it possible to reduce and optimize the evaluations for this cultrure. Biotechnology and molecular biology techniques are very useful to reduce the time and making more efficient the selection of new cultivars, as well as the development of high-throughput DNA sequencing technology in which genetic information can be obtained at lower costs with increases unprecedented accuracy. Meanwhile, advances in statistical analysis for quantitative genetics provide new methodologies for understanding the genetic basis of complex traits and will provide integrative information to increase our understanding of the balance of stress response of drought and latex yield.